MethylGPT
Foundation model for the human DNA methylome
Paper : “MethylGPT: a foundation model for the DNA methylome”
@article{ying2022methylgpt, title={MethylGPT: a foundation model for the DNA methylome}, author={Ying, Kejun and Song, Jinyeop and Cui, Haotian and Zhang, Yikun and Li, Siyuan and Chen, Xingyu and Liu, Hanna and Eames, Alec and McCartney, Daniel L and Marioni, Riccardo E and Poganik, Jesse R and Moqri, Mahdi and Wang, Bo and Gladyshev, Vadim N}, journal={}, year={2022} }
TLDR: We are building a foundation model for processing natural language representations of human methylation profiles, advancing research in biological aging and medicine.
Human DNA methylation data (methylome) is an important biomarker for aging and chronic diseases. Despite its significance, a unified and adaptable framework has yet to emerge, largely due to the absence of a “foundation model.” Foundation models have already proven essential for understanding the complexities of biology. For instance, in proteomics, models like ESM-2/ESM-3 and AlphaFold2/AlphaFold3 have achieved unprecedented accuracy in structure prediction and function annotation. In genomics, Enformer and Evo have demonstrated their ability to predict gene regulation and variant effects. Similarly, in single-cell biology, models such as Geneformer, scGPT, and scFoundation have enabled zero-shot cell-type classification and in-silico perturbation.
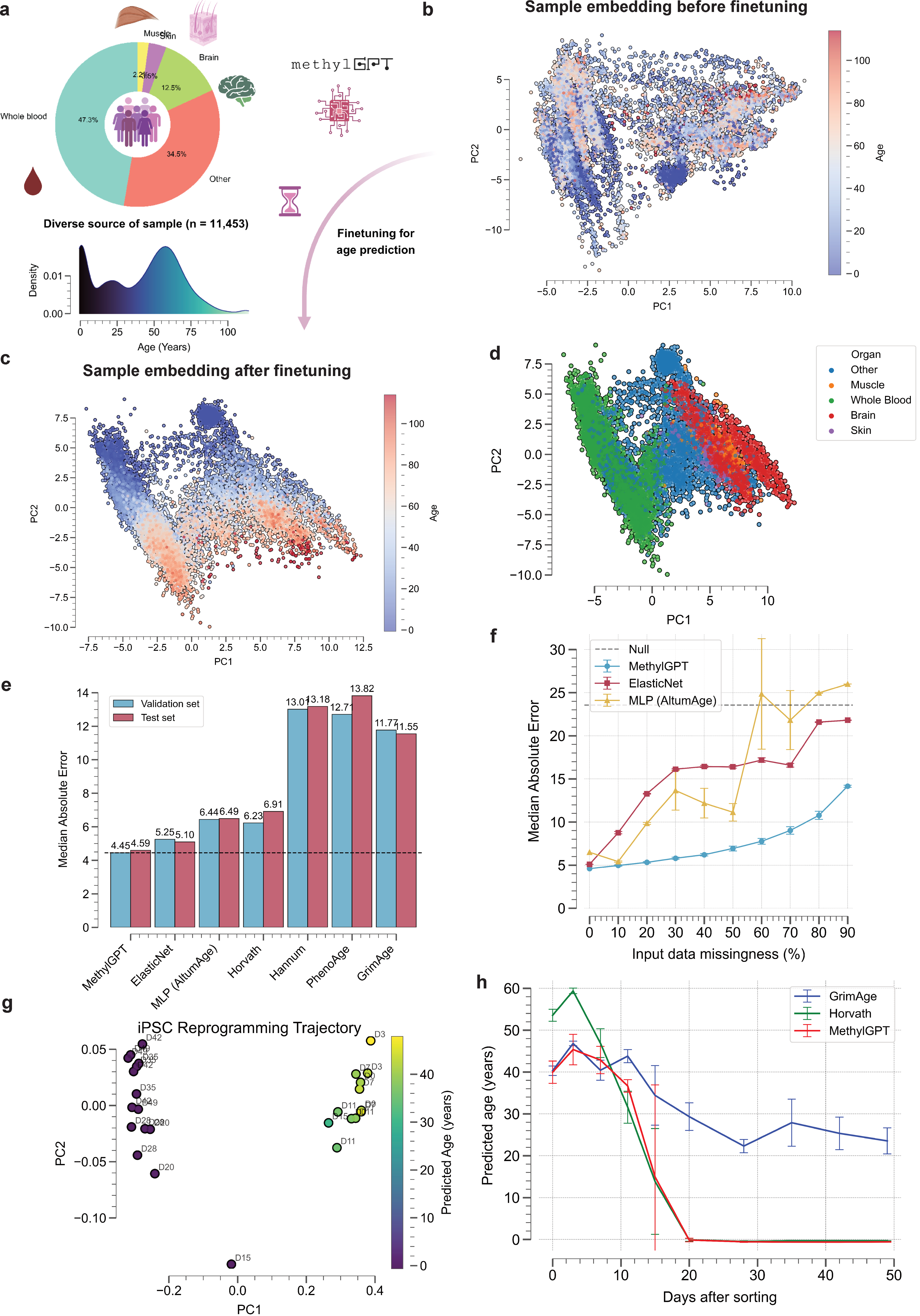
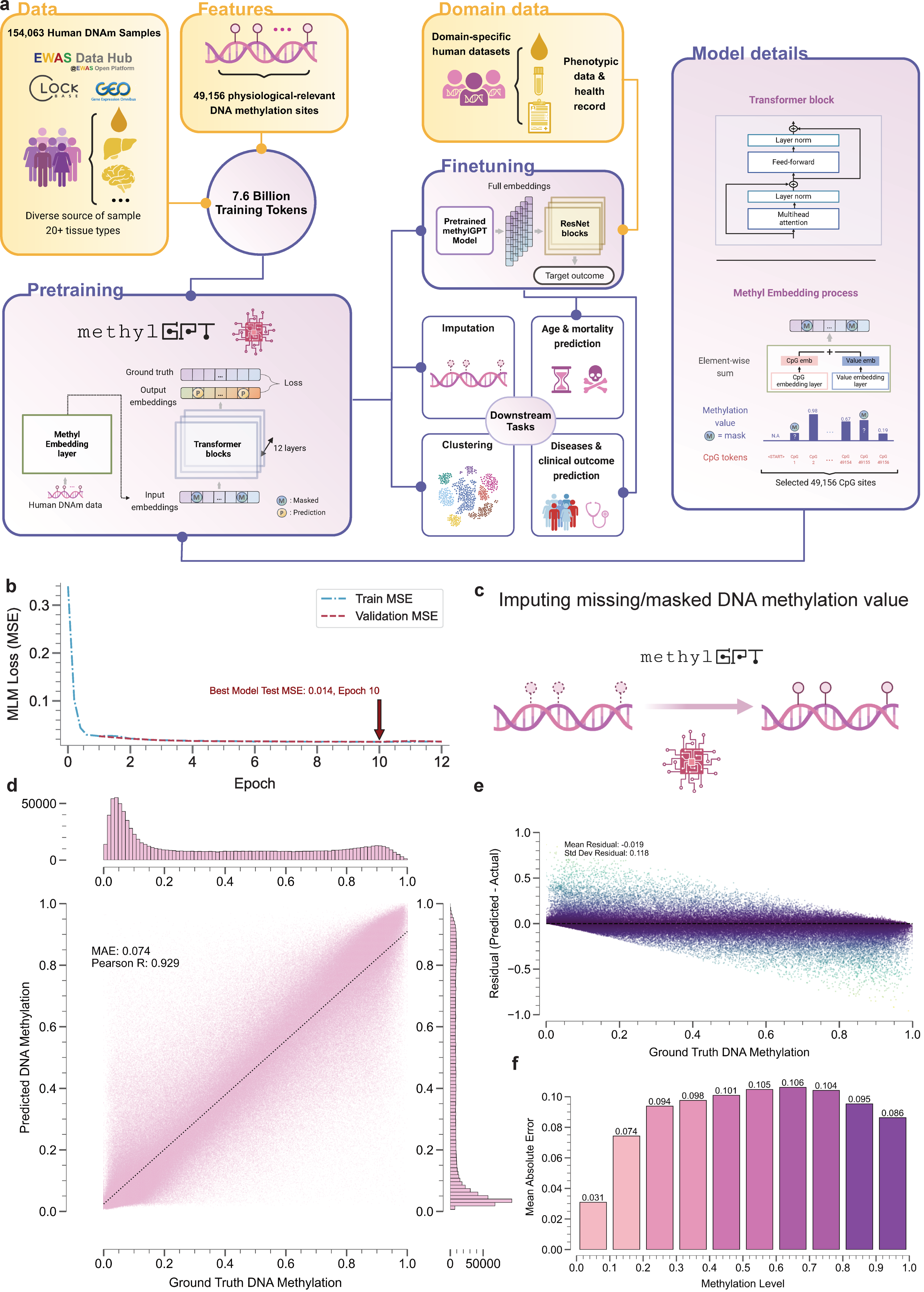
Therefore, our goal is to develop a foundational model, MethylGPT, specifically for human DNA methylation (DNAm) data, paving the way for future research. We curated approximately 300,000 DNAm samples from public sources, deduplicated them, and consolidated them into 154,063 unique human DNAm datasets. DNA methylation data vary in the number of CpG entries depending on the array platform used (e.g., Illumina 27k, Illumina 450k, and EPIC). To address these differences and ensure biological relevance, we focused on 49,156 CpG sites. In total, 7.6 billion training tokens were used for pretraining.
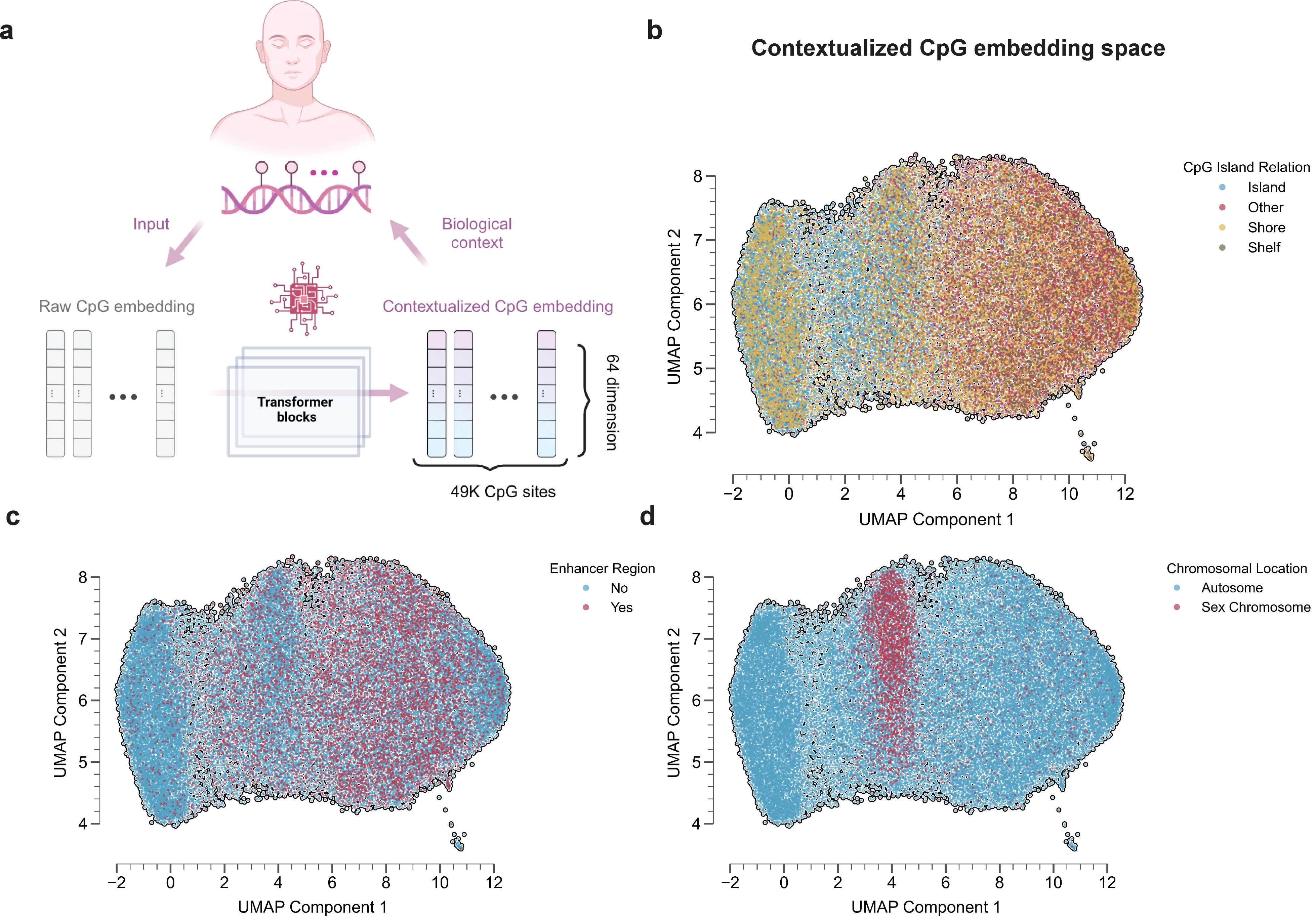
Our model architecture and training is specialized for DNAm data. First, we use the element-wise sum of the CpG value embedding and the CpG ID embedding. This allows information to be selectively masked without compromising the integrity of the sequence structure. Second, the training loss during the pretraining process includes both a Masked Language Modeling (MLM)-style approach and an Autoregressive Generative approach. Both approaches aim to optimize the prediction of methylation values to closely match the original values, even when some information is masked.
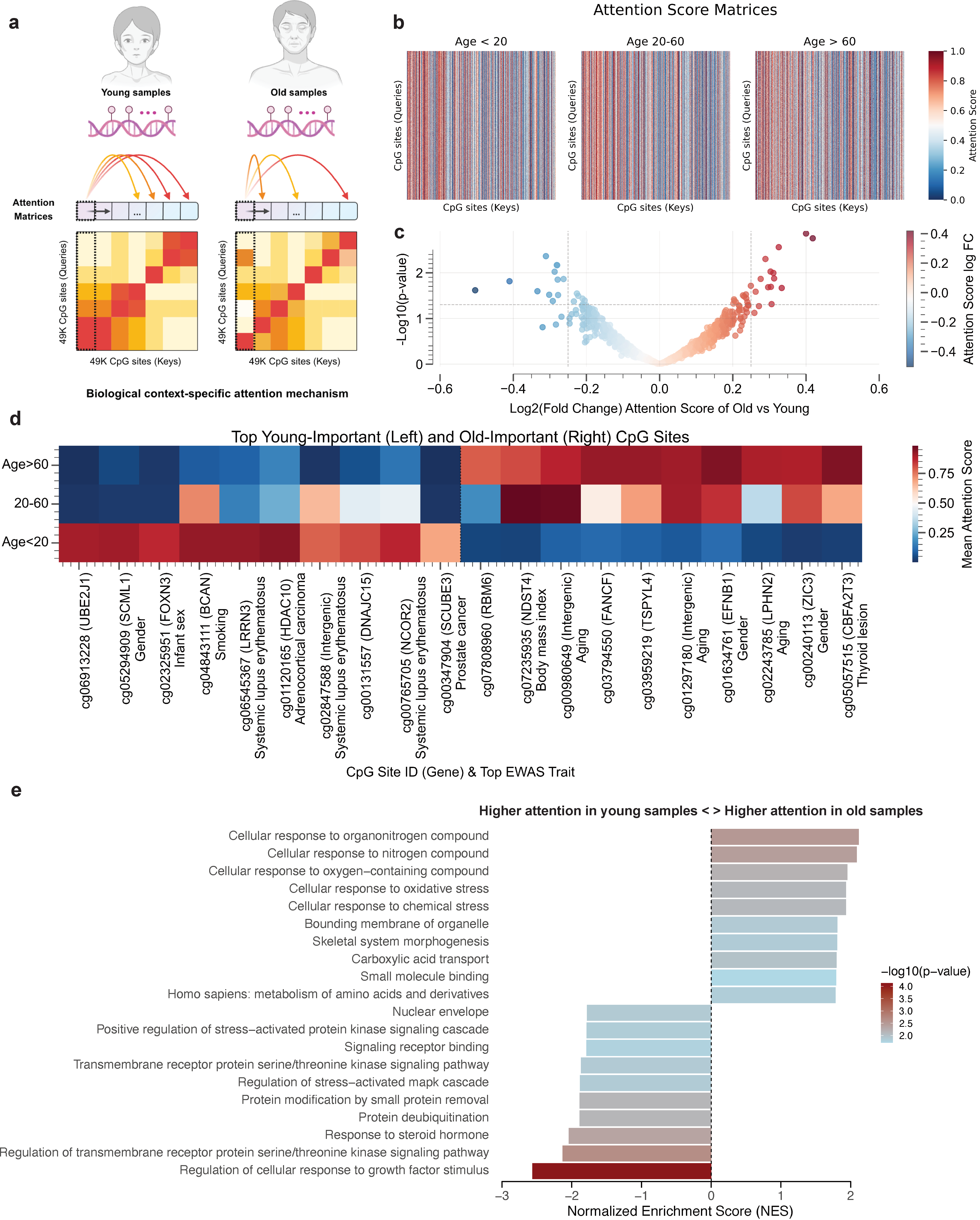
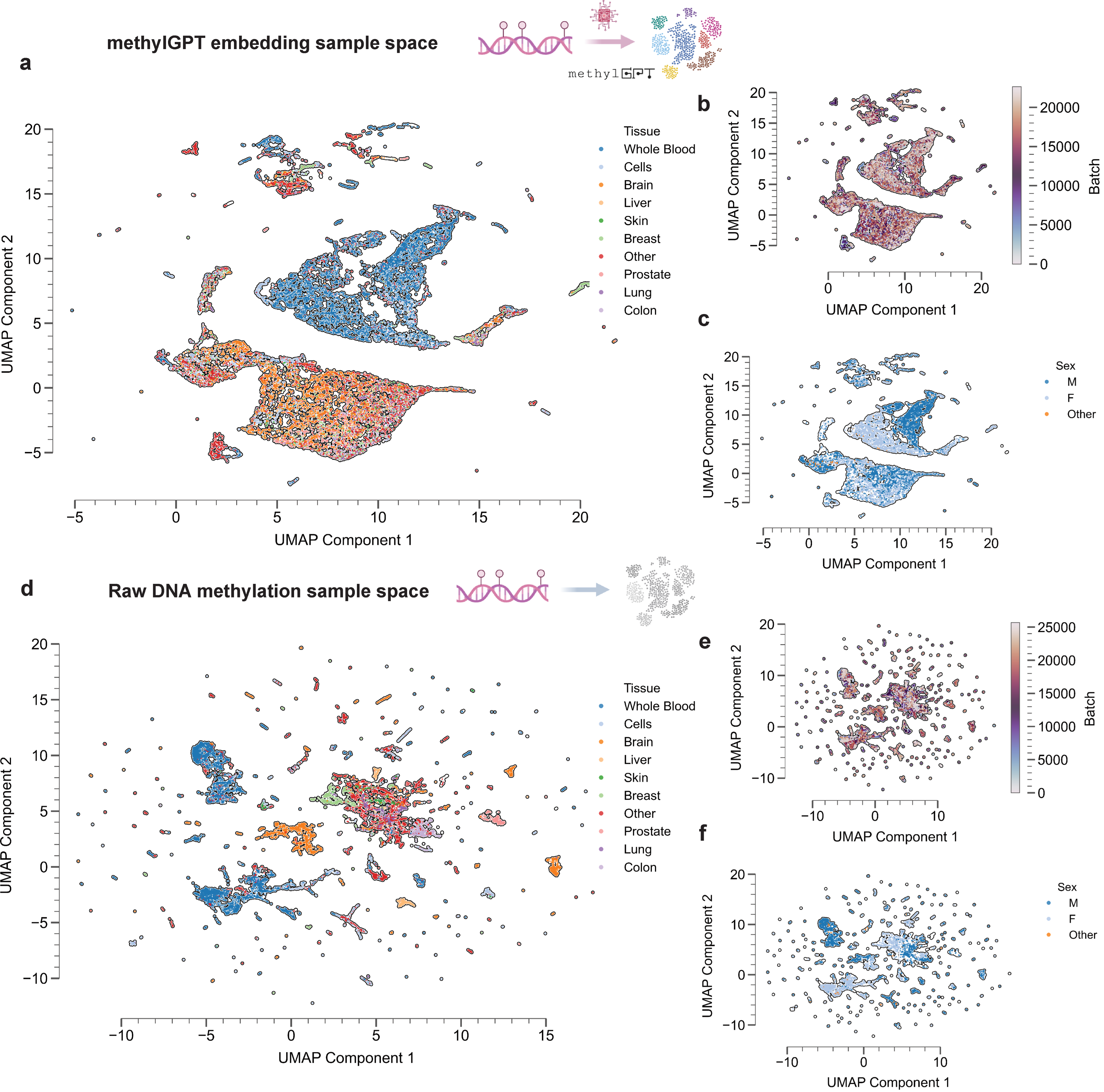
MethylGPT captures biologically meaningful sample-level features, such as tissue information, sex and disease tpyes compared than raw methylation data directly generated UMAP embeddings (Fig. 3d-f)
MethylGPT achieved superior accuracy( median absolute error (MedAE) of 4.45 ) in predicting biological age than other SOTA methods(ElasticNet [@zou2005], MLP (AltumAge) [@delimacamillo2022]).